
Newsroom
A research team led by Prof. WANG Jie from the Guangzhou Institutes of Biomedicine and Health of the Chinese Academy of Sciences, in collaboration with Prof. LI Yixue from Guangzhou National Laboratory, has developed a computational tool single-cell regulatory network inference (ScReNI). The method enables the identification of cell-specific regulatory networks and cell-enriched regulators, shedding light on how genes are regulated in individual cells across diverse biological processes. Their findings were recently published in Genomics, Proteomics & Bioinformatics.
Transcriptional regulatory networks are critical for shaping gene expression patterns in individual cells. The rise of single-cell sequencing technologies has driven advances in single-cell RNA sequencing (scRNA-seq) and single-cell ATAC sequencing (scATAC-seq), offering enhanced resolution to study gene regulation at the cellular level. These techniques reveal detailed transcriptomic and epigenomic landscapes, laying the groundwork for precise regulatory network inference. However, integrating scRNA-seq and scATAC-seq data—particularly when datasets are unpaired—remains a significant challenge.
To tackle this challenge, the team created ScReNI, a computational framework designed to infer cell-specific regulatory networks from either paired or unpaired scRNA-seq and scATAC-seq data. The tool first aligns the two data types in a shared analytical space, allowing integration even without matched cells. It then connects genes to regulatory peaks using genomic proximity analysis and transcription factor motif detection.
A key strength of ScReNI lies in its use of neighboring cell information to model local regulatory contexts. By combining k-nearest neighbors and random forest algorithms, the tool captures non-linear relationships between chromatin accessibility and gene expression, enabling the construction of personalized regulatory networks at single-cell resolution. Evaluations show ScReNI outperforms existing methods in predicting regulatory interactions and refining cell clusters, while also identifying key cell-specific regulatory factors—insights that deepen understanding of gene regulation mechanisms.
ScReNI advances single-cell genomics by enabling effective regulatory network inference and systematic identification of cell-specific regulators. It enhances the precision of gene regulatory predictions and captures dynamic regulatory mechanisms underlying complex biological processes such as development, differentiation, and disease progression.
The study was funded by the National Natural Science Foundation of China, the National Key Research and Development Program of China, and the Science and Technology Planning Project of Guangdong Province.
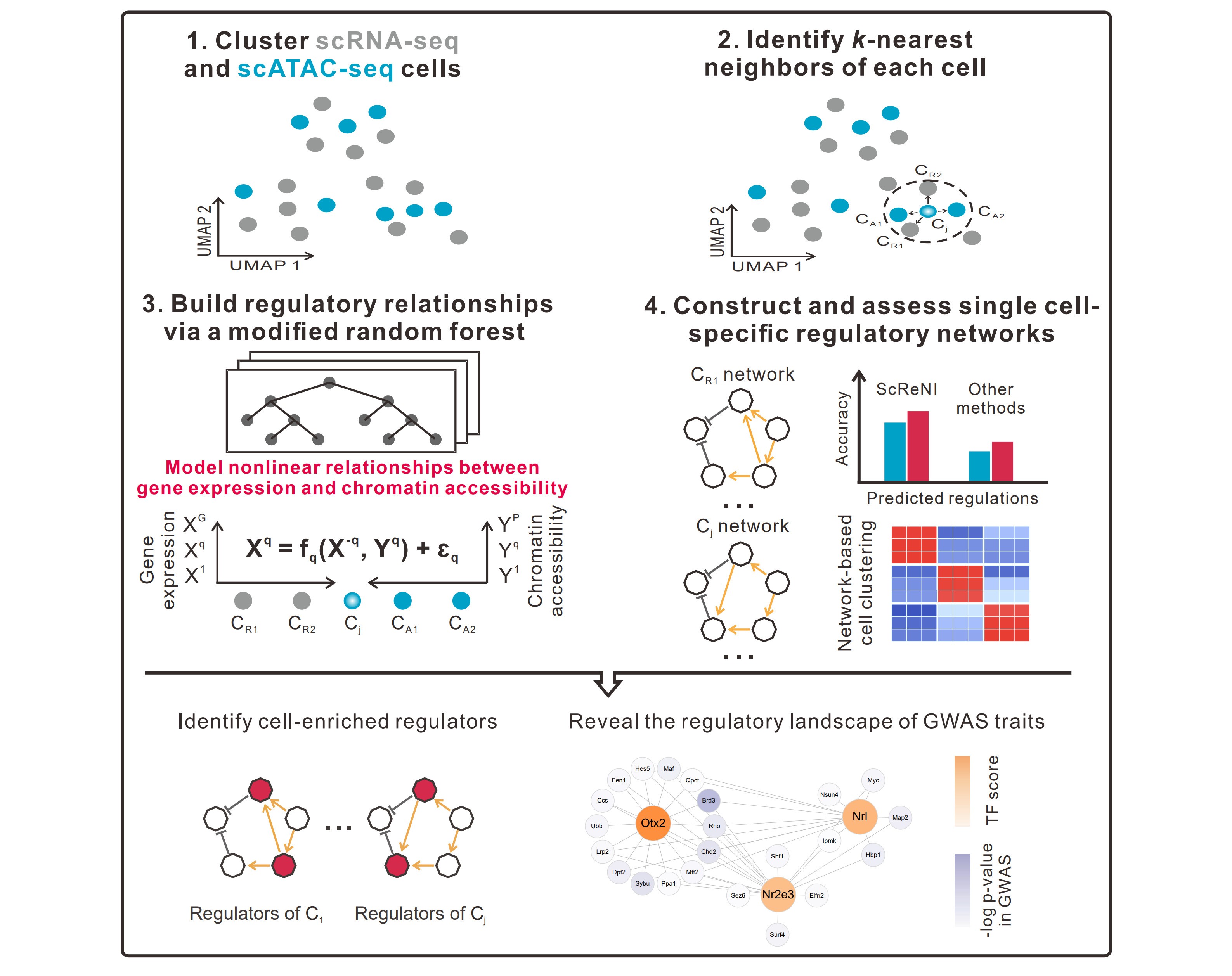
Overview of ScReNI (Image by Prof. WANG Jie's team)
