
tRNA is an adaptor molecule that enables the genetic code of nucleic acids to be converted to amino acids in protein. Consequently, tRNA is one of the most important molecules during protein synthesis. Post-transcriptional modifications are widely happened on tRNA, and those modifications are essential for tRNA to perform its cellular functions. Methylation of ribose moieties in tRNA is frequent, especially at position 32 where it is commonplace in all three domains of life. TrmJ proteins from the SPOUT methyltransferase superfamily are tRNA Xm32 modification enzymes that occur in bacteria and archaea. Unlike archaeal TrmJ, bacterial TrmJ require full-length tRNA molecules as substrates. It remains unknown how bacterial TrmJs recognize substrate tRNAs and specifically catalyze a 2′-O modification at ribose 32.
Dr. LIU Rujuan, PhD student LONG Tao and their colleagues, led by Prof. WANG Enduo from the Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences identified the tRNA initial binding mode of EcTrmJ by solving the crystal structures of EcTrmJ in complex with S-adenosyl-homocysteine, site directed mutations, ITC and electrophoretic mobility shift assays. Their results show that full-length EcTrmJ forms an unusual dimer in the asymmetric unit, with both the catalytic SPOUT domain and C-terminal extension forming separate dimeric associations. Furthermore, tRNA recognition by EcTrmJ involves the cooperative influences of conserved residues from both the SPOUT and extensional domains, and that this process is regulated by the flexible hinge region that connects these two domains.
This study entitled “tRNA recognition by a bacterial tRNA Xm32 modification enzyme from the SPOUT methyltransferase superfamily” was published online in the Nucleic Acids Research on July 21st, 2015.
The work is supported by grants from the Ministry of Science and Technology of China, the National Natural Science Foundation of China, the Chinese Academy of Sciences, and the Science and Technology Commission of Shanghai Municipality.
CONTACT:
WANG Enduo, Principal Investigator
Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences
Shanghai 200031, P. R. China
Phone: 86-21-54921241
E-mail: edwang@sibcb.ac.cn
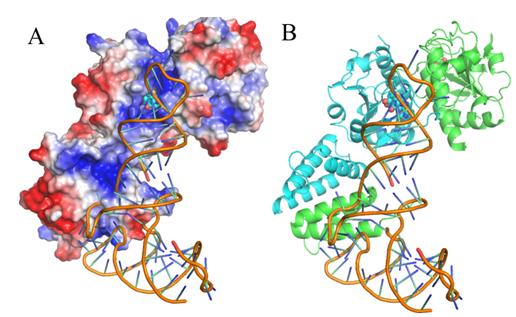
Fig. A proposed model of tRNA binding to EcTrmJ. (Image by Prof. WANG Enduo’s group)

86-10-68597521 (day)
86-10-68597289 (night)

86-10-68511095 (day)
86-10-68512458 (night)

cas_en@cas.cn

52 Sanlihe Rd., Xicheng District,
Beijing, China (100864)

